|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
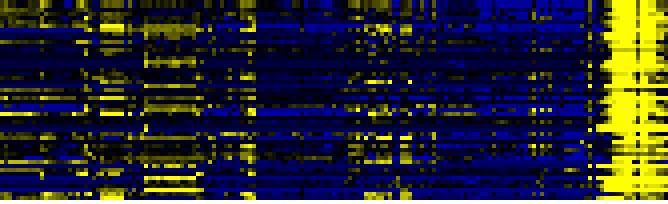
| Pathways co-expressed in the Organ and Tissue data set (with more than 10 annotation points) |
|
CYP704A1 / CYP704A2 (At2g44980 / At2g45510) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
6.3 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
4.4 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to organ heatmap |
|
|
|
|
|
|
|
| C-compound and carbohydrate metabolism |
FunCat |
43 |
0.000 |
9 |
0.001 |
|
|
|
| Pyruvate metabolism |
KEGG |
21 |
0.000 |
3 |
0.008 |
|
|
| glycolysis and gluconeogenesis |
FunCat |
20 |
0.000 |
4 |
0.010 |
|
|
| Intermediary Carbon Metabolism |
BioPath |
18 |
0.000 |
3 |
0.006 |
|
|
| IAA biosynthesis |
AraCyc |
18 |
0.000 |
2 |
0.000 |
|
|
| TCA cycle variation VII |
AraCyc |
18 |
0.000 |
3 |
0.010 |
|
|
| Phenylpropanoid pathway |
LitPath |
18 |
0.000 |
4 |
0.001 |
|
|
| Phenylpropanoid Metabolism |
BioPath |
16 |
0.000 |
3 |
0.008 |
|
|
| glyoxylate cycle |
AraCyc |
16 |
0.000 |
2 |
0.003 |
|
|
| TCA cycle variation VIII |
AraCyc |
16 |
0.000 |
2 |
0.022 |
|
|
| Glyoxylate and dicarboxylate metabolism |
KEGG |
16 |
0.000 |
2 |
0.004 |
|
|
| Miscellaneous acyl lipid metabolism |
AcylLipid |
16 |
0.000 |
4 |
0.100 |
|
|
| core phenylpropanoid metabolism |
BioPath |
14 |
0.000 |
2 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| Glutathione metabolism |
KEGG |
14 |
0.000 |
2 |
0.011 |
|
|
|
|
|
|
|
|
|
|
|
| fermentation |
FunCat |
13 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| plant / fungal specific systemic sensing and response |
FunCat |
13 |
0.000 |
2 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| plant hormonal regulation |
FunCat |
13 |
0.000 |
2 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| Benzoate degradation via CoA ligation |
KEGG |
13 |
0.000 |
3 |
0.017 |
|
|
|
|
|
|
|
|
|
|
|
| Glycolysis / Gluconeogenesis |
KEGG |
13 |
0.000 |
2 |
0.055 |
|
|
|
|
|
|
|
|
|
|
|
| Tryptophan metabolism |
KEGG |
13 |
0.000 |
3 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| lignin biosynthesis |
AraCyc |
12 |
0.000 |
2 |
0.016 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in the Stress data set ( with more than 6 annotation points) |
|
CYP704A1 / CYP704A2 (At2g44980 / At2g45510) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
3.1 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
1.6 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to stress heatmap |
|
|
|
|
|
|
|
| Biosynthesis of Amino Acids and Derivatives |
BioPath |
22 |
0.000 |
5 |
0.000 |

|
| Glutathione metabolism |
BioPath |
22 |
0.000 |
5 |
0.000 |
| Fatty acid metabolism |
KEGG |
19 |
0.000 |
2 |
0.001 |
| Glutathione metabolism |
KEGG |
12 |
0.000 |
2 |
0.002 |
| Gluconeogenesis from lipids in seeds |
BioPath |
10 |
0.000 |
1 |
0.031 |
| Degradation of storage lipids and straight fatty acids |
AcylLipid |
10 |
0.000 |
1 |
0.006 |
| C-compound and carbohydrate metabolism |
FunCat |
9 |
0.004 |
1 |
0.172 |
|
|
|
|
|
|
|
|
|
|
|
| fermentation |
FunCat |
9 |
0.000 |
1 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| 1- and 2-Methylnaphthalene degradation |
KEGG |
9 |
0.000 |
1 |
0.003 |
|
|
|
|
|
|
|
|
|
|
|
| beta-Alanine metabolism |
KEGG |
9 |
0.000 |
1 |
0.007 |
|
|
|
|
|
|
|
|
|
|
|
| Bile acid biosynthesis |
KEGG |
9 |
0.000 |
1 |
0.004 |
|
|
|
|
|
|
|
|
|
|
|
| Glycerolipid metabolism |
KEGG |
9 |
0.000 |
1 |
0.019 |
|
|
|
|
|
|
|
|
|
|
|
| Glycolysis / Gluconeogenesis |
KEGG |
9 |
0.002 |
1 |
0.086 |
|
|
|
|
|
|
|
|
|
|
|
| Methane metabolism |
KEGG |
9 |
0.000 |
1 |
0.094 |
|
|
|
|
|
|
|
|
|
|
|
| Nucleotide Metabolism |
KEGG |
9 |
0.001 |
1 |
0.085 |
|
|
|
|
|
|
|
|
|
|
|
| Pantothenate and CoA biosynthesis |
KEGG |
9 |
0.000 |
1 |
0.007 |
|
|
|
|
|
|
|
|
|
|
|
| Pyrimidine metabolism |
KEGG |
9 |
0.000 |
1 |
0.027 |
|
|
|
|
|
|
|
|
|
|
|
| Pyruvate metabolism |
KEGG |
9 |
0.002 |
1 |
0.077 |
|
|
|
|
|
|
|
|
|
|
|
| Tyrosine metabolism |
KEGG |
9 |
0.000 |
1 |
0.027 |
|
|
|
|
|
|
|
|
|
|
|
| biosynthesis of phenylpropanoids |
FunCat |
8 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| biosynthesis of secondary products derived from L-phenylalanine and L-tyrosine |
FunCat |
8 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| detoxification |
FunCat |
8 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|







