Co-Expression Analysis of:
CYPedia Home
Institut de Biologie Moléculaire des Plantes (Home)
CYP706A2 (At4g22710)
save all data as Tab Delimited Table
Pathways co-expressed in all 4 data sets (with more than 6 annotation points each)
Find below a list of pathways that are co-expressed with the bait. First a list of pathways is given that are co-expressed in all data sets. Lists for each individual dataset are shown underneath. Depending on the number of co-expressed pathways only the top scoring pathways are given; all data can be saved as text using the link above.

Pathway
Source
Sum of scores
Sum of genes
Glutathione metabolism
BioPath
56
8
Glutathione metabolism
KEGG
56
8
To the right of each table a thumbnail of the actual co-expression heatmap is given. Klick on the link to see the heatmap containing all co-expressed genes.
For more information on how these pathway maps were generated please read the methods page
Pathways co-expressed in the Organ and Tissue data set (with more than 9 annotation points)
CYP706A1 (At4g22690)
max. difference between log2-ratios:
7.5
max. difference between log2-ratios excluding lowest and highest 5%:
6.4
Pathway
Source
Scores of Genes
p[Score]
No. of Genes
p[genes]
Link to organ heatmap
Biosynthesis of Amino Acids and Derivatives
BioPath
46
0.000
6
0.000
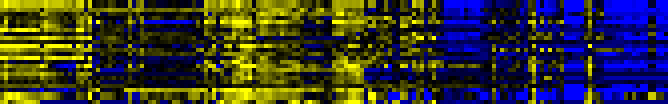
Glutathione metabolism
BioPath
22
0.000
3
0.001
toxin catabolism
TAIR-GO
22
0.000
3
0.001
Glutathione metabolism
KEGG
22
0.000
3
0.000
Stilbene, coumarine and lignin biosynthesis
KEGG
19
0.000
3
0.006
Methionin/SAM/ethylene metabolism from cysteine and aspartate
BioPath
16
0.000
2
0.001
intracellular signalling
FunCat
13
0.000
3
0.000
Benzoate degradation via CoA ligation
KEGG
13
0.000
3
0.004
Inositol phosphate metabolism
KEGG
13
0.000
3
0.005
Nicotinate and nicotinamide metabolism
KEGG
13
0.000
3
0.003
lignin biosynthesis
AraCyc
12
0.000
2
0.000
Phenylpropanoid pathway
LitPath
12
0.000
2
0.002
core phenylpropanoid metabolism
BioPath
10
0.000
1
0.033
aging
TAIR-GO
10
0.000
1
0.002
lignin biosynthesis
TAIR-GO
10
0.000
1
0.003
response to stress
TAIR-GO
10
0.000
1
0.001
response to water deprivation
TAIR-GO
10
0.000
1
0.000
triacylglycerol degradation
AraCyc
10
0.000
1
0.006
biogenesis of cell wall
FunCat
10
0.000
1
0.037
Miscellaneous acyl lipid metabolism
AcylLipid
10
0.000
1
0.000
Pathways co-expressed in the Stress data set ( with more than 6 annotation points)
CYP706A1 (At4g22690)
max. difference between log2-ratios:
6.4
max. difference between log2-ratios excluding lowest and highest 5%:
2.7
Pathway
Source
Scores of Genes
p[Score]
No. of Genes
p[genes]
Link to stress heatmap
Biosynthesis of Amino Acids and Derivatives
BioPath
12
0.000
2
0.004

Glutathione metabolism
BioPath
12
0.000
2
0.000
sulfate assimilation
TAIR-GO
10
0.000
1
0.000
dissimilatory sulfate reduction
AraCyc
10
0.000
1
0.000
sulfate assimilation III
AraCyc
10
0.000
1
0.002
biogenesis of chloroplast
FunCat
10
0.000
1
0.007
nitrogen and sulfur utilization
FunCat
10
0.000
1
0.000
Purine metabolism
KEGG
10
0.000
1
0.022
Selenoamino acid metabolism
KEGG
10
0.000
1
0.006
Sulfur metabolism
KEGG
10
0.000
1
0.003
nucleotide metabolism
TAIR-GO
9
0.000
1
0.000
de novo biosynthesis of purine nucleotides I
AraCyc
9
0.000
1
0.015
Pathways co-expressed in the Hormone etc. data set (with more than 6 annotation points)
CYP706A1 (At4g22690)
max. difference between log2-ratios:
3.4
max. difference between log2-ratios excluding lowest and highest 5%:
1.2
Pathway
Source
Scores of Genes
p[Score]
No. of Genes
p[genes]
Link to hormones etc. heatmap
Miscellaneous acyl lipid metabolism
AcylLipid
12
0.000
2
0.019

Gluconeogenesis from lipids in seeds
BioPath
10
0.000
1
0.006
fatty acid biosynthesis
TAIR-GO
10
0.000
1
0.002
fatty acid oxidation pathway
AraCyc
10
0.000
1
0.000
octane oxidation
AraCyc
10
0.000
1
0.000
degradation
FunCat
10
0.000
1
0.002
degradation of lipids, fatty acids and isoprenoids
FunCat
10
0.000
1
0.000
Carbon fixation
KEGG
10
0.000
2
0.005
Fatty acid metabolism
KEGG
10
0.000
1
0.014
auxin mediated signaling pathway
TAIR-GO
9
0.000
1
0.000
calcium ion binding
TAIR-GO
9
0.000
1
0.000
Mo-molybdopterin cofactor biosynthesis
TAIR-GO
9
0.000
1
0.000
molybdenum ion binding
TAIR-GO
9
0.000
1
0.000
response to absence of light
TAIR-GO
9
0.000
1
0.000
response to mechanical stimulus
TAIR-GO
9
0.000
1
0.000
response to temperature
TAIR-GO
9
0.000
1
0.000
thigmotropism
TAIR-GO
9
0.000
1
0.000
biosynthesis of vitamins, cofactors, and prosthetic groups
FunCat
9
0.000
1
0.001
Folate biosynthesis
KEGG
9
0.000
1
0.003
Phosphatidylinositol signaling system
KEGG
9
0.000
1
0.014
Signal Transduction
KEGG
9
0.000
1
0.015
Pathways co-expressed in the Mutant data set (with more than 10 annotation points)
CYP706A1 (At4g22690)
max. difference between log2-ratios:
4.8
max. difference between log2-ratios excluding lowest and highest 5%:
2.2
Pathway
Source
Scores of Genes
p[Score]
No. of Genes
p[genes]
Link to mutants heatmap
Benzoate degradation via CoA ligation
KEGG
37
0.000
8
0.000
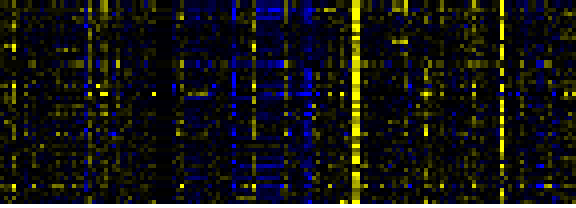
Inositol phosphate metabolism
KEGG
37
0.000
8
0.000
Nicotinate and nicotinamide metabolism
KEGG
37
0.000
8
0.000
Phenylpropanoid Metabolism
BioPath
32
0.000
5
0.009
Miscellaneous acyl lipid metabolism
AcylLipid
24
0.000
5
0.002
Biosynthesis of Amino Acids and Derivatives
BioPath
22
0.004
3
0.104
Aromatic amino acid (Phe, Tyr, Trp) metabolism
BioPath
20
0.000
3
0.003
Phenylpropanoid pathway
LitPath
20
0.000
4
0.005
N-terminal protein myristoylation
TAIR-GO
18
0.000
2
0.000
degradation
FunCat
18
0.000
2
0.002
tryptophan biosynthesis
AraCyc
16
0.000
3
0.000
Glutamate metabolism
KEGG
16
0.000
2
0.014
tryptophan biosynthesis
TAIR-GO
14
0.000
2
0.001
Shikimate pathway
LitPath
14
0.000
2
0.022
Trp biosyntesis
LitPath
14
0.000
2
0.002
Glutathione metabolism
BioPath
12
0.002
2
0.038
nucleotide metabolism
FunCat
12
0.000
2
0.000
trehalose biosynthesis I
AraCyc
11
0.000
2
0.026
trehalose biosynthesis II
AraCyc
11
0.000
2
0.010
trehalose biosynthesis III
AraCyc
11
0.000
2
0.002
energy
FunCat
11
0.000
2
0.000
metabolism of energy reserves (e.g. glycogen, trehalose)
FunCat
11
0.000
2
0.000
page created by Juergen Ehlting
01/02/06