|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
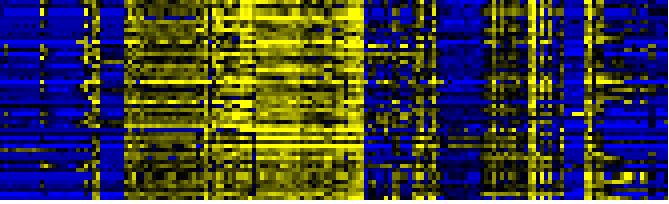
| Pathways co-expressed in the Organ and Tissue data set (with more than 20 annotation points) |
|
CYP72A8(At3g14620) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
7.9 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
5.4 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to organ heatmap |
|
|
|
|
|
|
|
| Phenylpropanoid Metabolism |
BioPath |
44 |
0.010 |
6 |
0.239 |
|
|
|
| Plastidial Isoprenoids (Chlorophylls, Carotenoids, Tocopherols, Plastoquinone, Phylloquinone) |
BioPath |
40 |
0.000 |
6 |
0.010 |
|
|
| transport |
FunCat |
40 |
0.000 |
5 |
0.000 |
|
|
| Lipid signaling |
AcylLipid |
38 |
0.000 |
8 |
0.017 |
|
|
| photorespiration |
AraCyc |
32 |
0.000 |
6 |
0.000 |
|
|
| Miscellaneous acyl lipid metabolism |
AcylLipid |
32 |
0.000 |
6 |
0.183 |
|
|
| Carotenoid and abscisic acid metabolism |
LitPath |
32 |
0.000 |
4 |
0.000 |
|
|
| Glucosyltransferases for benzoic acids |
BioPath |
30 |
0.000 |
3 |
0.000 |
|
|
| biogenesis of chloroplast |
FunCat |
30 |
0.000 |
4 |
0.025 |
|
|
| transport facilitation |
FunCat |
30 |
0.000 |
4 |
0.000 |
|
|
| Ion channels |
KEGG |
30 |
0.000 |
7 |
0.000 |
|
|
| Ligand-Receptor Interaction |
KEGG |
30 |
0.000 |
7 |
0.000 |
|
|
| Chloroplastic protein turnover |
BioPath |
28 |
0.000 |
4 |
0.000 |
|
|
| hypersensitive response |
TAIR-GO |
28 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| photorespiration |
TAIR-GO |
28 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Carotenoid biosynthesis |
BioPath |
26 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| carotenoid biosynthesis |
AraCyc |
26 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| energy |
FunCat |
26 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| carotenid biosynthesis |
LitPath |
26 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Carbon fixation |
KEGG |
25 |
0.000 |
3 |
0.022 |
|
|
|
|
|
|
|
|
|
|
|
| additional photosystem II components |
BioPath |
24 |
0.000 |
3 |
0.026 |
|
|
|
|
|
|
|
|
|
|
|
| jasmonic acid biosynthesis |
TAIR-GO |
24 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| jasmonic acid biosynthesis |
AraCyc |
24 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| photosynthesis |
FunCat |
24 |
0.000 |
4 |
0.007 |
|
|
|
|
|
|
|
|
|
|
|
| Biosynthesis of steroids |
KEGG |
24 |
0.000 |
3 |
0.007 |
|
|
|
|
|
|
|
|
|
|
|
| ATP-dependent proteolysis |
TAIR-GO |
22 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| calmodulin binding |
TAIR-GO |
22 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| glycine biosynthesis I |
AraCyc |
22 |
0.000 |
4 |
0.005 |
|
|
|
|
|
|
|
|
|
|
|
| superpathway of serine and glycine biosynthesis II |
AraCyc |
22 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| degradation |
FunCat |
21 |
0.000 |
3 |
0.005 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
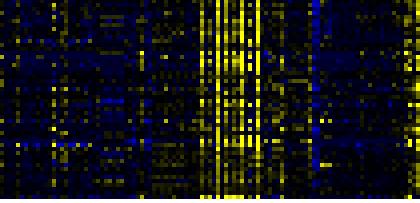
| Pathways co-expressed in the Hormone etc. data set (with more than 20 annotation points) |
|
CYP72A8(At3g14620) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
4.8 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
2.0 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to hormones etc. heatmap |
|
|
|
|
|
|
|
| Phenylpropanoid Metabolism |
BioPath |
105 |
0.000 |
14 |
0.000 |
|
|
|
|
|
|
| Biosynthesis of Amino Acids and Derivatives |
BioPath |
60 |
0.000 |
7 |
0.024 |
|
|
|
|
|
| Glutathione metabolism |
BioPath |
60 |
0.000 |
7 |
0.000 |
|
|
|
|
|
| tryptophan biosynthesis |
TAIR-GO |
58 |
0.000 |
8 |
0.000 |
|
|
|
|
|
| tryptophan biosynthesis |
AraCyc |
58 |
0.000 |
8 |
0.000 |
|
|
|
|
|
| Shikimate pathway |
LitPath |
58 |
0.000 |
8 |
0.000 |
|
|
|
|
|
| Trp biosyntesis |
LitPath |
58 |
0.000 |
8 |
0.000 |
|
|
|
|
|
| Aromatic amino acid (Phe, Tyr, Trp) metabolism |
BioPath |
54 |
0.000 |
8 |
0.000 |
|
|
|
|
|
| response to pathogenic bacteria |
TAIR-GO |
54 |
0.000 |
8 |
0.000 |
|
|
|
|
|
| sulfate assimilation III |
AraCyc |
44 |
0.000 |
6 |
0.000 |
|
|
|
|
|
| Phenylalanine, tyrosine and tryptophan biosynthesis |
KEGG |
36 |
0.000 |
6 |
0.000 |
|
|
|
|
|
| cysteine biosynthesis I |
AraCyc |
35 |
0.000 |
5 |
0.000 |
|
|
|
|
|
| Glucosyltransferases for benzoic acids |
BioPath |
30 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Intermediary Carbon Metabolism |
BioPath |
30 |
0.042 |
6 |
0.049 |
|
|
|
|
|
|
|
|
|
|
|
| toxin catabolism |
TAIR-GO |
26 |
0.000 |
9 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| nitrogen and sulfur metabolism |
FunCat |
26 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Gluconeogenesis from lipids in seeds |
BioPath |
24 |
0.000 |
3 |
0.056 |
|
|
|
|
|
|
|
|
|
|
|
| lipid, fatty acid and isoprenoid degradation |
FunCat |
22 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Glutathione metabolism |
KEGG |
22 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|







