|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in both data sets with co-expressed genes |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Sum of scores |
Sum of genes |
|
Find below a list of pathways that are co-expressed with the bait. First a list of pathways is given that are co-expressed in all four data sets. Lists for each individual dataset are shown underneath. To the right of each table a thumbnail of the actual co-expression heatmap is given. Klick on the link to see the heatmap containing all co-expressed genes. |
|
|
|
|
|
|
|
|
|
| Miscellaneous acyl lipid metabolism |
AcylLipid |
98 |
21 |
|
|
|
|
|
|
|
|
|
|
| prenyl diphosphate (GPP,FPP, GGPP) biosynthesis |
LitPath |
56 |
13 |
|
|
|
|
|
|
|
|
|
|
| sexual reproduction |
TAIR-GO |
40 |
5 |
|
|
|
|
|
|
|
|
|
|
| Fatty acid elongation and wax and cutin metabolism |
AcylLipid |
37 |
7 |
|
|
|
|
|
|
|
|
|
|
| Isoprenoid Biosynthesis in the Cytosol and in Mitochondria |
BioPath |
34 |
3 |
|
For more information on how these pathway maps were generated please read the methods page |
|
|
|
|
|
|
|
|
|
| Biosynthesis of prenyl diphosphates |
BioPath |
26 |
2 |
|
|
|
|
|
|
|
|
|
|
| fatty acid metabolism |
TAIR-GO |
22 |
3 |
|

|
|
|
|
|
|
|
|
|
|
|
| long-chain fatty acid metabolism |
TAIR-GO |
22 |
3 |
|
|
|
|
|
|
|
|
|
|
|
| very-long-chain fatty acid metabolism |
TAIR-GO |
22 |
3 |
|
|
|
|
|
|
|
|
|
|
|
| lignin biosynthesis |
AraCyc |
20 |
5 |
|
|
|
|
|
|
|
|
|
|
|
| Phenylpropanoid pathway |
LitPath |
20 |
5 |
|
|
|
|
|
|
|
|
|
|
|
|
| cuticle biosynthesis |
TAIR-GO |
16 |
2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
| suberin biosynthesis |
AraCyc |
14 |
3 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in the Organ and Tissue data set |
|
CYP98A9 (At1g74550) |
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
5.33 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
2.02 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to organ heatmap |
|
|
|
|
|
|
|
| Miscellaneous acyl lipid metabolism |
AcylLipid |
44 |
0.000 |
16 |
0.000 |
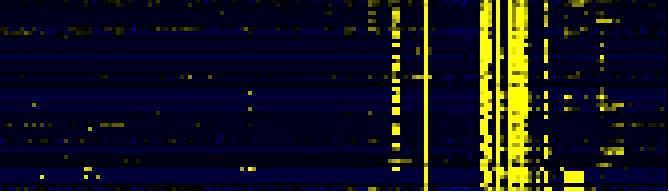
|
|
|
|
| sexual reproduction |
TAIR-GO |
20 |
0.000 |
5 |
0.000 |
|
|
|
| Fatty acid elongation and wax and cutin metabolism |
AcylLipid |
15 |
0.001 |
5 |
0.046 |
|
|
|
| Biosynthesis of prenyl diphosphates |
BioPath |
14 |
0.000 |
3 |
0.000 |
|
|
|
| Isoprenoid Biosynthesis in the Cytosol and in Mitochondria |
BioPath |
14 |
0.000 |
3 |
0.000 |
|
|
|
| prenyl diphosphate (GPP,FPP, GGPP) biosynthesis |
LitPath |
10 |
0.000 |
1 |
0.012 |
|
|
|
| lignin biosynthesis |
AraCyc |
9 |
0.000 |
4 |
0.000 |
|
|
|
| Phenylpropanoid pathway |
LitPath |
9 |
0.000 |
4 |
0.001 |
|
|
|
| cuticle biosynthesis |
TAIR-GO |
8 |
0.000 |
2 |
0.000 |
|
|
|
| fatty acid metabolism |
TAIR-GO |
8 |
0.000 |
2 |
0.001 |
|
|
|
| long-chain fatty acid metabolism |
TAIR-GO |
8 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| very-long-chain fatty acid metabolism |
TAIR-GO |
8 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| suberin biosynthesis |
AraCyc |
7 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in the Mutant data set |
|
CYP98A9 (At1g74550) |
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
5.07 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
1.32 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
|
|
|
|
|
|
|
|
|
|
|
| Cell Wall Carbohydrate Metabolism |
BioPath |
93 |
0.000 |
24 |
0.000 |
Link to mutants heatmap |
|
|
|
|
|
|
|
| pectin metabolism |
BioPath |
77 |
0.000 |
21 |
0.000 |
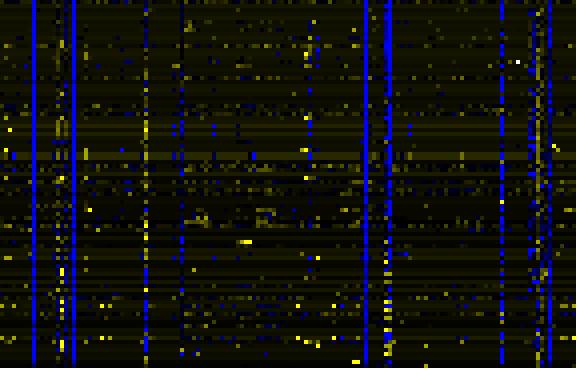
|
|
|
|
|
| Miscellaneous acyl lipid metabolism |
AcylLipid |
54 |
0.000 |
21 |
0.000 |
|
|
|
|
| C-compound and carbohydrate metabolism |
FunCat |
46 |
0.000 |
13 |
0.008 |
|
|
|
|
| Starch and sucrose metabolism |
KEGG |
33 |
0.000 |
6 |
0.000 |
|
|
|
|
| biogenesis of cell wall |
FunCat |
26 |
0.000 |
9 |
0.000 |
|
|
|
|
| Glycan Biosynthesis and Metabolism |
KEGG |
26 |
0.000 |
4 |
0.002 |
|
|
|
|
| Fatty acid elongation and wax and cutin metabolism |
AcylLipid |
22 |
0.000 |
7 |
0.043 |
|
|
|
|
| Isoprenoid Biosynthesis in the Cytosol and in Mitochondria |
BioPath |
20 |
0.000 |
3 |
0.097 |
|
|
|
|
| sexual reproduction |
TAIR-GO |
20 |
0.000 |
5 |
0.000 |
|
|
|
|
| Glutamate/glutamine from nitrogen fixation |
BioPath |
14 |
0.000 |
2 |
0.005 |
|
|
|
|
| fatty acid metabolism |
TAIR-GO |
14 |
0.000 |
3 |
0.000 |
|
|
|
|
| long-chain fatty acid metabolism |
TAIR-GO |
14 |
0.000 |
3 |
0.000 |
|
|
|
|
| very-long-chain fatty acid metabolism |
TAIR-GO |
14 |
0.000 |
3 |
0.000 |
|
|
|
|
| lactose degradation IV |
AraCyc |
14 |
0.000 |
4 |
0.000 |
|
|
|
|
| assimilation of ammonia, metabolism of the glutamate group |
FunCat |
14 |
0.000 |
2 |
0.017 |
|
|
|
|
| nitrogen and sulfur metabolism |
FunCat |
14 |
0.000 |
2 |
0.014 |
|
|
|
|
| Glutamate metabolism |
KEGG |
14 |
0.000 |
2 |
0.023 |
|
|
|
|
| Glycerolipid metabolism |
KEGG |
14 |
0.000 |
2 |
0.011 |
|
|
|
|
| Nitrogen metabolism |
KEGG |
14 |
0.000 |
2 |
0.010 |
|
|
|
|
| Biosynthesis of prenyl diphosphates |
BioPath |
12 |
0.009 |
2 |
0.123 |
|
|
|
|
|
|
|
|
|
|
|
| fatty acid biosynthesis -- initial steps |
AraCyc |
12 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| triacylglycerol degradation |
AraCyc |
12 |
0.000 |
5 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| Galactose metabolism |
KEGG |
12 |
0.000 |
2 |
0.010 |
|
|
|
|
|
|
|
|
|
|
|
| lignin biosynthesis |
AraCyc |
11 |
0.001 |
5 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| isoprenoid biosynthesis |
FunCat |
11 |
0.038 |
3 |
0.075 |
|
|
|
|
|
|
|
|
|
|
|
| lipid, fatty acid and isoprenoid biosynthesis |
FunCat |
11 |
0.016 |
3 |
0.049 |
|
|
|
|
|
|
|
|
|
|
|
| Phenylpropanoid pathway |
LitPath |
11 |
0.019 |
5 |
0.002 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|





