|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in the Hormone etc. data set (with more than 12 annotation points) |
|
CYP78A10 (At1g74110) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
4.0 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
0.0 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to hormones etc. heatmap |
|
|
|
|
|
|
|
| Lipid signaling |
AcylLipid |
48 |
0.000 |
13 |
0.004 |
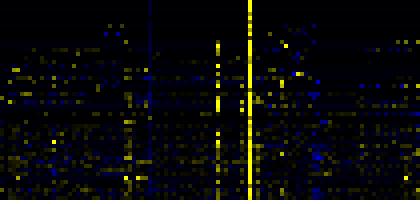
|
|
|
|
|
|
| Inositol phosphate metabolism |
KEGG |
46 |
0.000 |
14 |
0.000 |
|
|
|
|
|
| Biosynthesis of Amino Acids and Derivatives |
BioPath |
42 |
0.002 |
8 |
0.024 |
|
|
|
|
|
| Leaf Glycerolipid Biosynthesis |
BioPath |
42 |
0.015 |
7 |
0.074 |
|
|
|
|
|
| Benzoate degradation via CoA ligation |
KEGG |
42 |
0.000 |
13 |
0.000 |
|
|
|
|
|
| Nicotinate and nicotinamide metabolism |
KEGG |
42 |
0.000 |
13 |
0.000 |
|
|
|
|
|
| Plastidial Isoprenoids (Chlorophylls, Carotenoids, Tocopherols, Plastoquinone, Phylloquinone) |
BioPath |
40 |
0.000 |
4 |
0.059 |
|
|
|
|
|
| secondary metabolism |
FunCat |
36 |
0.000 |
7 |
0.000 |
|
|
|
|
|
| Methionin/SAM/ethylene metabolism from cysteine and aspartate |
BioPath |
34 |
0.000 |
6 |
0.000 |
|
|
|
|
|
| Chlorophyll biosynthesis and breakdown |
BioPath |
30 |
0.000 |
3 |
0.009 |
|
|
|
|
|
| Leaf Glycerolipid Biosynthesis in Plastid |
BioPath |
30 |
0.000 |
3 |
0.056 |
|
|
|
|
|
| Propanoate metabolism |
KEGG |
30 |
0.000 |
5 |
0.001 |
|
|
|
|
|
| Synthesis of membrane lipids in plastids |
AcylLipid |
30 |
0.000 |
3 |
0.018 |
|
|
|
|
|
| chlorophyll and phytochromobilin metabolism |
LitPath |
30 |
0.000 |
3 |
0.028 |
|
|
|
|
|
|
|
|
|
|
|
| triacylglycerol degradation |
AraCyc |
29.5 |
0.000 |
8 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Phenylpropanoid pathway |
LitPath |
28 |
0.001 |
5 |
0.059 |
|
|
|
|
|
|
|
|
|
|
|
| Glycerolipid metabolism |
KEGG |
26 |
0.000 |
3 |
0.033 |
|
|
|
|
|
|
|
|
|
|
|
| Glycan Biosynthesis and Metabolism |
KEGG |
25 |
0.000 |
7 |
0.002 |
|
|
|
|
|
|
|
|
|
|
|
| ethylene biosynthesis |
TAIR-GO |
24 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| biosynthesis of proto- and siroheme |
AraCyc |
24 |
0.000 |
3 |
0.024 |
|
|
|
|
|
|
|
|
|
|
|
| cellulose biosynthesis |
BioPath |
20 |
0.000 |
3 |
0.027 |
|
|
|
|
|
|
|
|
|
|
|
| cellular response to phosphate starvation |
TAIR-GO |
20 |
0.000 |
2 |
0.002 |
|
|
|
|
|
|
|
|
|
|
|
| galactolipid biosynthesis |
TAIR-GO |
20 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| lipid biosynthesis |
TAIR-GO |
20 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| glycosylglyceride biosynthesis |
AraCyc |
20 |
0.000 |
2 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| Fructose and mannose metabolism |
KEGG |
20 |
0.000 |
3 |
0.090 |
|
|
|
|
|
|
|
|
|
|
|
| Ion channels |
KEGG |
20 |
0.000 |
10 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Ligand-Receptor Interaction |
KEGG |
20 |
0.000 |
10 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Porphyrin and chlorophyll metabolism |
KEGG |
20 |
0.000 |
2 |
0.077 |
|
|
|
|
|
|
|
|
|
|
|
| phytochromobilin biosynthesis |
LitPath |
20 |
0.000 |
2 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| systemic acquired resistance |
TAIR-GO |
19.5 |
0.000 |
3 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| intracellular signalling |
FunCat |
19 |
0.000 |
5 |
0.003 |
|
|
|
|
|
|
|
|
|
|
|
| defense response |
TAIR-GO |
16.5 |
0.000 |
3 |
0.005 |
|
|
|
|
|
|
|
|
|
|
|
| hemicellulose biosynthesis |
BioPath |
16 |
0.000 |
2 |
0.007 |
|
|
|
|
|
|
|
|
|
|
|
| fructose degradation (anaerobic) |
AraCyc |
16 |
0.000 |
4 |
0.027 |
|
|
|
|
|
|
|
|
|
|
|
| lignin biosynthesis |
AraCyc |
16 |
0.000 |
4 |
0.006 |
|
|
|
|
|
|
|
|
|
|
|
| phospholipid biosynthesis II |
AraCyc |
16 |
0.000 |
3 |
0.003 |
|
|
|
|
|
|
|
|
|
|
|
| sorbitol fermentation |
AraCyc |
16 |
0.001 |
4 |
0.032 |
|
|
|
|
|
|
|
|
|
|
|
| plant / fungal specific systemic sensing and response |
FunCat |
16 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| plant hormonal regulation |
FunCat |
16 |
0.000 |
4 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| de novo biosynthesis of purine nucleotides I |
AraCyc |
15 |
0.005 |
4 |
0.085 |
|
|
|
|
|
|
|
|
|
|
|
| lipases pathway |
AraCyc |
15 |
0.000 |
3 |
0.005 |
|
|
|
|
|
|
|
|
|
|
|
| Aromatic amino acid (Phe, Tyr, Trp) metabolism |
BioPath |
14 |
0.027 |
2 |
0.166 |
|
|
|
|
|
|
|
|
|
|
|
| core phenylpropanoid metabolism |
BioPath |
14 |
0.009 |
2 |
0.107 |
|
|
|
|
|
|
|
|
|
|
|
| chlorophyll biosynthesis |
AraCyc |
14 |
0.003 |
2 |
0.165 |
|
|
|
|
|
|
|
|
|
|
|
| biogenesis of chloroplast |
FunCat |
14 |
0.019 |
2 |
0.216 |
|
|
|
|
|
|
|
|
|
|
|
| lipid, fatty acid and isoprenoid degradation |
FunCat |
14 |
0.000 |
2 |
0.033 |
|
|
|
|
|
|
|
|
|
|
|
| Glycerophospholipid metabolism |
KEGG |
14 |
0.000 |
2 |
0.052 |
|
|
|
|
|
|
|
|
|
|
|
| defense response to pathogenic bacteria, incompatible interaction |
TAIR-GO |
12.5 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| response to pathogenic bacteria |
TAIR-GO |
12.5 |
0.000 |
2 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|



