|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in the Organ and Tissue data set (with more than 6 annotation points) |
|
CYP96A9 (At4g39480) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
8.0 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
4.9 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to organ heatmap |
|
|
|
|
|
|
|
| mono-/sesqui-/di-terpene biosynthesis |
LitPath |
40 |
0.000 |
4 |
0.000 |
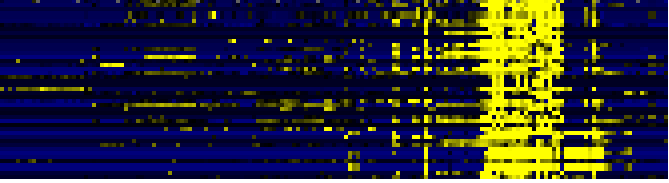
|
|
|
| terpenoid metabolism |
LitPath |
40 |
0.000 |
4 |
0.001 |
|
|
| Cell Wall Carbohydrate Metabolism |
BioPath |
30.5 |
0.000 |
5 |
0.040 |
|
|
| pectin metabolism |
BioPath |
30.5 |
0.000 |
5 |
0.001 |
|
|
| Fatty acid elongation and wax and cutin metabolism |
AcylLipid |
24 |
0.000 |
4 |
0.007 |
|
|
| Starch and sucrose metabolism |
KEGG |
20 |
0.000 |
2 |
0.000 |
|
|
| fatty acid metabolism |
TAIR-GO |
18 |
0.000 |
2 |
0.000 |
|
|
| Synthesis of membrane lipids in endomembrane system |
AcylLipid |
11 |
0.012 |
3 |
0.022 |
|
|
| Gluconeogenesis from lipids in seeds |
BioPath |
10 |
0.000 |
1 |
0.100 |
|
|
| monoterpenoid biosynthesis |
TAIR-GO |
10 |
0.000 |
1 |
0.000 |
|
|
| pentacyclic triterpenoid biosynthesis |
TAIR-GO |
10 |
0.000 |
1 |
0.001 |
|
|
| fatty acid oxidation pathway |
AraCyc |
10 |
0.000 |
1 |
0.002 |
|
|
| monoterpene biosynthesis |
AraCyc |
10 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| octane oxidation |
AraCyc |
10 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| plant monoterpene biosynthesis |
AraCyc |
10 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| Biosynthesis of steroids |
KEGG |
10 |
0.000 |
1 |
0.004 |
|
|
|
|
|
|
|
|
|
|
|
| monoterpene biosynthesis |
LitPath |
10 |
0.000 |
1 |
0.005 |
|
|
|
|
|
|
|
|
|
|
|
| sequiterpene biosynthesis |
LitPath |
10 |
0.000 |
1 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| triterpene biosynthesis |
LitPath |
10 |
0.000 |
1 |
0.021 |
|
|
|
|
|
|
|
|
|
|
|
| cuticle biosynthesis |
TAIR-GO |
7 |
0.000 |
1 |
0.002 |
|
|
|
|
|
|
|
|
|
|
|
| wax biosynthesis |
TAIR-GO |
7 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| epicuticular wax biosynthesis |
AraCyc |
7 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Pathways co-expressed in the Mutant data set (with more than 6 annotation points) |
|
CYP96A9 (At4g39480) |
|
|
|
|
|
|
|
|
| max. difference between log2-ratios: |
6.0 |
|
|
|
|
|
|
|
|
|
|
|
|
| max. difference between log2-ratios excluding lowest and highest 5%: |
2.3 |
|
|
|
|
|
|
|
|
|
|
|
|
| Pathway |
Source |
Scores of Genes |
p[Score] |
No. of Genes |
p[genes] |
Link to mutants heatmap |
|
|
|
|
|
|
|
| fatty acid beta oxidation complex |
BioPath |
7 |
0.000 |
1 |
0.002 |

|
|
|
|
| Gluconeogenesis from lipids in seeds |
BioPath |
7 |
0.000 |
1 |
0.016 |
|
|
|
| fatty acid beta-oxidation |
TAIR-GO |
7 |
0.000 |
1 |
0.000 |
|
|
|
| fatty acid oxidation pathway |
AraCyc |
7 |
0.000 |
1 |
0.001 |
|
|
|
| isoleucine degradation I |
AraCyc |
7 |
0.000 |
1 |
0.002 |
|
|
|
|
|
|
|
|
|
|
|
| isoleucine degradation III |
AraCyc |
7 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| leucine degradation I |
AraCyc |
7 |
0.000 |
1 |
0.002 |
|
|
|
|
|
|
|
|
|
|
|
| leucine degradation II |
AraCyc |
7 |
0.000 |
1 |
0.000 |
|
|
|
|
|
|
|
|
|
|
|
| valine degradation I |
AraCyc |
7 |
0.000 |
1 |
0.003 |
|
|
|
|
|
|
|
|
|
|
|
| valine degradation II |
AraCyc |
7 |
0.000 |
1 |
0.001 |
|
|
|
|
|
|
|
|
|
|
|
| Degradation of storage lipids and straight fatty acids |
AcylLipid |
7 |
0.000 |
1 |
0.006 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|



